Test Patterns: Unlocking the Bacterial Code of Recurrent and Antibiotic-Tolerant Ear Infections
By Elisa Ludwig
Any parent of young children knows all too well the misery of middle ear infections — many of which are recurrent and some of which can tolerate antibiotic treatment. But what if there were a way to predict such outcomes based on bacterial analysis? Better still, what if there were drugs specifically designed to help target these tough cases?
For some time, Mell has been intrigued by the human bacterial pathogen nontypeable Haemophilus influenzae (or NTHi), which causes ear infections (otitis media) in children, and lung infections for those with chronic respiratory illness. Mell has been particularly interested in a process called “natural transformation,” in which cells actively transport environmental DNA through their cell membranes where homologous molecules can be incorporated into chromosomes. This allows NTHi and other “naturally competent bacteria” to acquire genes from their relatives, which can spread antibiotic resistance genes and other pathogenesis traits.
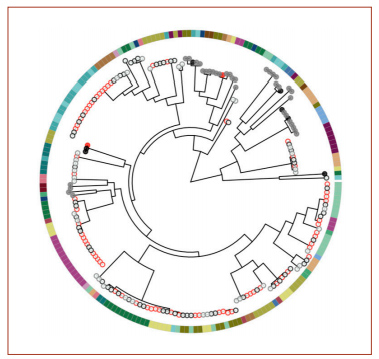
A phylogenetic tree from the genome analysis. Red circles indicate samples collected from a sick child’s ear; black circles are from a sick child’s nose; gray circles are from a healthy child’s nose. Filled circles indicate the presence of two genes, potE and speF. The colors around the outside indicate different children.
Joshua Chang Mell, PhD, assistant professor in the Department of Microbiology & Immunology, is tackling these questions in his study “Detecting and Disrupting Biofilms in Recurrent Ear Infections,” for which he was awarded a 2020 Hartwell Foundation Grant.
Most recently, he embarked on genome-wide association studies to connect natural genetic variation in NTHi bacteria with the risk of pediatric middle ear infections. This involves collaboration with PIs in Drexel’s Center for Advanced Microbial Processing (CAMP), including Garth D. Ehrlich, PhD, professor, Departments of Microbiology & Immunology, and Otolaryngology-Head & Neck Surgery, and Joris Beld, PhD, assistant professor, Department of Microbiology & Immunology, along with Michael Pichichero, MD, director of the Rochester General Hospital Research Institute, which holds a vast archive of tens of thousands of bacteria specimens, dating back more than 15 years, from healthy children’s nasal passages and from the middle ears of children with otitis media.
“By sequencing and then comparing the highly diverse genomes of more than 200 NTHi isolates collected from healthy and disease states, we identified several bacterial genes whose presence or absence predicted isolation from heath or disease,” Mell says. “These were nearly all involved in pH homeostasis and nitrogen metabolism. This turned out to be quite interesting clinically, since it’s recently been shown that the fluid from infected middle ears is often alkaline (high pH), and we found bacterial genes whose products modify extracellular pH.”
Mell was surprised by the results, which found less evidence for virulence genes, but rather evidence for the opposite.
“We had really set out to find genes whose presence in the bacterium contributes to pathogenesis during middle ear disease. Instead, out of six candidate genes, five of the ‘hits’ were associations between disease and gene absence,” he says. “On the one hand, this offers much less obvious routes toward translational research. A bona fide virulence gene could become the target for a new drug or therapy, but it’s hard to design a drug against something that isn’t there. On the other hand, the literature shows us many instances of bacteria that normally don’t cause disease that can become pathogenic upon losing gene functions.”
These findings were the basis for intended follow-up mechanistic studies with candidate genes and the team’s proposal to the Hartwell Foundation.
“Our main hypothesis is that bacteria that are more likely to form biofilms in the middle ear environment are also more likely to cause recurrent ear infections, a major health problem for many young children,” he says. “We are now working to examine the relationship between pH changes and biofilm formation, a trait that allows bacteria to persist within infection, in spite of antibiotic treatment. Notably, the machinery in NTHi required for biofilms is also required for natural competence. Our preliminary data suggest that different strains are highly variable in their biofilm formation and in the environmental conditions that trigger it.”
The first part of this study involves testing new designer biofilm inhibitor compounds developed by recent Drexel graduate Donald J. Hall, PhD chemistry ’20, CAMP lab manager Jaroslaw Krol, PhD, and Ehrlich, along with Frank Ji, PhD, professor, Department of Chemistry in Drexel’s College of Arts and Sciences, on diverse bacteria strains in diverse conditions. Mell says that these drugs may synergize with antibiotics to eliminate persistent bacteria that reseed infections.
NTHi biofilms grown in neutral pH. Live cells are green, while dead ones are red. The double-mutation of
potE and speF caused more robust growth (right) than the wild type (left).
“We’re taking advantage of the fact that we know a lot about how diverse these bacteria are, how they have these incredibly large genetic differences between them. And we know that some genes that we’ve identified are involved in the process of infection. So, we have some hints about how to conduct these efforts and how to study these bacteria in test tubes to see how the drugs work.”
The second piece of the research consists of measuring global gene expression in the samples, using RNA sequencing across diverse strains and conditions, to design a new diagnostic for use on specimens that have been taken from children with middle ear infections. Here, Mell’s team is searching for signatures that indicate whether the infection is likely to be a recurring one — in particular, a pattern of genes whose expression presages biofilm formation.
“With this new diagnostic, we will directly test clinical specimens from children with sporadic or recurrent ear infections to see whether signatures of biofilm formation predict recurrent disease,” he says. “This could act as a new diagnostic to identify children at high risk for repeated infections because of the bacteria they host.”
The diagnostic, specifically designed to assess NTHi, could be used in other NTHi-related conditions, such as chronic obstructive pulmonary disease (COPD) in elderly patients. The methodology and pipeline used to develop NTHi diagnostics could readily be applied to other bacterial pathogens, Mell says.
The ramifications of this research — the idea that the evolutionary genomics can tell us more about how to diagnose and treat illness in the future — are exciting to Mell, who was trained as a basic researcher in genetics.
“Before, my work was very indirectly applicable to anything translational. But now, I work at a medical school on a biomedically relevant bacterial pathogen. It wasn’t until this transition that I’d really thought much or learned much about bacterial pathogenesis at all,” he says. “I’m trying to let the bacteria tell us what matters to them with minimal preconceptions. By watching how bacteria adapt on their hosts during recurring or chronic infections, we can hopefully find new ways to undermine them that people hadn’t thought about.”
The biofilm research represents the interdisciplinary approach to biology that Mell and others are taking at CAMP. Aided by advances in nucleic acid sequencing technology and metabolite profiling, researchers are mining microbial genomes, metagenomes and hologenomes in the search for better target molecules and new antimicrobial compounds.
“Projects that would’ve cost millions of dollars just 10 to 15 years ago can now be done by a regular microbiology lab that has a little bit of computational skillset. Much of biology has become split between experimentalists with a lot of biology domain knowledge and computational biologists/bioinformaticists who can handle heavy data analysis and statistics but don’t know a lot of biology. There are real barriers to communication between these people because of training, even if they work in the same lab,” Mell says. “So, at CAMP we try to cross-train ourselves, and our staff members and students, to act at the interface between wet-lab experiments and dry-lab analysis. We also not only acknowledge the profound genetic diversity within organisms—both host and pathogen — but also accommodate and use this diversity in our experimental design.”
For example, he says, MD/PhD student E. Ari Gordon is developing a novel analysis framework for looking at bacterial gene expression measurements that don’t rely on a single “reference” strain, whose genomes may greatly differ from others. Far more sensitive and accurate than typical methods, it’s being used to look at the commonalities between diverse NTHi biofilms.
Mell and his team will likely be submitting results from the initial studies by summer, and he anticipates completing the phenotypic assays around the diverse strains and conditions within a year, the gene expression analysis within two years, and the testing on clinical specimens relatively soon after that.
“I’d like to think that we are at least on the cutting edge, if not outright innovative in our work,” Mell says. “It’s an exciting time to be a microbiologist, and especially to be a microbial geneticist/genomicist.”
Back to Top